Non interactive Multimedia
1st Place: Urban Absorption in New Bedford, MA (1888-2010)
Justin Hollander, Assistant Professor, Urban and Environmental Policy, School of Arts and Sciences
Rui Guo, Graduate Student, Urban and Environmental Policy, School of Arts and Sciences
Abstract
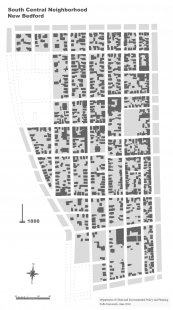
As part of Dr. Hollander’s ongoing research into the ways that cities manage population decline, the attached illustration is an effort to better visualize and communicate the way that one neighborhood in New Bedford, Massachusetts has physically changed during a period of major population loss. The file is a moving gif, intended to be used on a research project website in the coming months.
By digitizing Sanborn Fire Insurance Maps over the last one-hundred years Dr. Hollander and his research assistant, Gui Rui has made major progress in understanding a pattern of decline seemingly imperceptible to the naked eye. By layering figure-ground drawings of several New Bedford neighborhoods we can see where the city shrunk. How the physical form of the city accommodated a loss of 30, 105 persons in 89 years, what the disappearance of tens of thousands of housing units looks like.
Cities shrink for many reasons, but in the case of New Bedford, job losses are the biggest factor. As employment fell, first with the Great Depression, then due to the devastation caused by the 1938 Hurricane, people left. With fewer jobs, there are fewer people, but oddly houses tend to linger. In fact, housing is quite durable, it hardly disappears when a city begins to depopulate. But somehow, despite their study construction, tens of thousands of New Bedford’s houses have slowly disappeared and vacant land appears in its place. The attached illustration helps to frame key research questions that Dr. Hollander’s work is probing: But what becomes of the land when buildings are gone, how is the city remade in the wake of depopulation? How do designers and planners make sense of the uncertainty surrounding such change?
2nd Place: XROMM Pony
Bronwen A. Childs, Graduate Student, Comparative Medical Sciences, Cummings School of Veterinary Medicine
This text will be replaced
My video submission is designed to showcase the research I am doing in the field of equine kinematics (motion studies). Horses are incredible athletes superbly designed for speed and agility with their long legs designed with the heavy muscle mass kept close to the body and long tendons and ligaments transferring the forces that control the lighter lower limb. The delicate balance of speed and stability rests on the proper functioning and efficient biomechanics of the network of joints. When the functional apparatus fails the result is often a catastrophic breakdown injury, a fatal misstep for most sport horses. The most commonly injured joint is the metacarpophalangeal joint or “fetlock” and the goal of my research is to create a precise and accurate reconstruction of the three-dimensional (3D) motion of the fetlock in order to bolster the body of knowledge upon which therapies and supportive bandaging can be developed. I am using a technique called X-ray Reconstruction of Moving Morphology or “XROMM” to create the 3D model. The technique involves capturing the live motion with x-ray video from two orthogonal angles and then using that motion to animate 3D bone models derived from a computed tomography (CT) scan of the same limb. In order to facilitate matching the bones with the motion defined by the x-ray video, radio-opaque (dark in x-rays) markers were fixed to the surface of the bones that make up the joint of interest. The video sequences are digitized by tracking the markers in each frame and then bone models from a CT scan (with the same markers labeled) are paired with the marker locations in the moving x-ray and animated to follow that motion. The 3D bone models provide morphologically accurate reconstructions of the joint and the motion is measured with a precision of 0.09mm, making XROMM the most precise method currently available for motion analysis. XROMM has been employed in other species including humans, dogs, and fish, but this study marks the first use of XROMM in the horse.
3rd Place: Heapviz: A Programmer’s Tool for Data Structure Visualization
Visit entry’s web site
Edward Aftandilian, Graduate Student, Computer Science, School of Arts and Sciences
Sean Kelley, Undergraduate Student, School of Arts and Sciences
Connor Gramazio, Undergraduate Student, School of Arts and Sciences
Nathan Ricci, Graduate Student, Computer Science Dept, School of Arts and Sciences
Sara Su, Visiting Assistant Professor, Computer Science, School of Arts and Sciences